Technion researchers develop a technology for encoding, retrieving, and rapidly reading data stored in DNA.
Researchers from the Henry and Marilyn Taub Faculty of Computer Science have developed an AI-based method that accelerates DNA-based data retrieval by three orders of magnitude while significantly improving accuracy. The research team included Ph.D. student Omer Sabary, Dr. Daniella Bar-Lev, Dr. Itai Orr, Prof. Eitan Yaakobi, and Prof. Tuvi Etzion.

DNA data storage is an emerging field that leverages DNA as a platform for storing information. DNA offers significant advantages as a storage medium, including:
- Long-Term Preservation: In 2013, researchers in Denmark successfully extracted DNA from a horse bone dating back 700,000 years. In 2021, an international team recovered DNA from mammoths that lived over a million years ago. By contrast, magnetic disks used in data centers have lifespans measured in years or, at best, a few decades. This highlights DNA’s potential for long-term storage.
- Energy and Cost Efficiency: The “cloud” that powers most of today’s computing services relies on data centers that consume approximately 3% of global electricity and emit around 2% of total carbon emissions. With the exponential growth of data, the environmental impact of existing technologies is expected to increase significantly.
- Unmatched Data Density: DNA storage offers data density up to 100 million times greater than traditional digital storage. This means that a volume currently holding one megabyte could theoretically store up to 100 terabytes using DNA.
DNA is a molecule composed of a sequence of organic compounds called nucleotides. These nucleotides are classified into four types, represented by the letters A, C, G, and T. Unlike traditional computing, where data is encoded using only two digits (0 and 1), DNA storage is based on sequences of four letters, dramatically increasing the number of possible combinations.
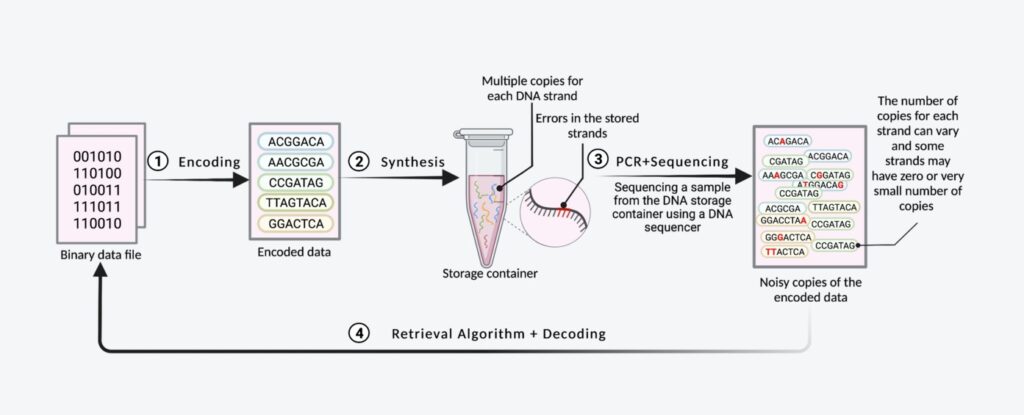
To write (store) data in this technology, DNA synthesis is required – creating DNA molecules based on the sequences encoding the information. To read the stored data, DNA sequencing is necessary.

Credit: Rami Shlush, Technion Spokesperson’s Office
Challenges in DNA Data Storage
Developing DNA-based storage technology presents several technological challenges:
- Both synthesis and sequencing are lengthy and error-prone processes, introducing deletion, insertion, and substitution errors
- Due to the limitations of the synthesis process, multiple copies of each DNA molecule encoding the data are produced. These copies are stored together, unordered, in a storage container
- During sequencing, many erroneous copies of these molecules are retrieved – most containing errors, while some disappear entirely
DNAformer: AI-Powered Data Retrieval
The current research presents a comprehensive computational solution for retrieving and correcting errors in complex DNA-based storage systems. Using advanced algorithms and encoding techniques, the researchers have demonstrated that their solution reduces data retrieval and reading time from several days to just 10 minutes.
The Technion-developed method, DNAformer, is based on a transformer model trained on simulated data (generated using a simulator, which was also developed at the Technion) to reconstruct accurate DNA sequences from erroneous copies. The method also includes a custom error-correction code tailored for DNA, ensuring robust data integrity.
Additionally, an extra safety margin mechanism detects particularly noisy DNA sequences (unwanted signals or errors that occur during the sequencing process, which can interfere with the accurate interpretation of the data) and applies powerful algorithmic tools to handle them efficiently. At the end of the process, the data is converted back into digital information.
Breakthrough Performance
The new method enables the reading of 100 megabytes of data at a speed 3,200 times faster than the most accurate existing method – without any loss of accuracy. Compared to previously known fast methods, DNAformer also improves accuracy by up to 40% while significantly reducing processing time. This was demonstrated on a 3.1-megabyte dataset, which included:
- A color still image
- A 24-second audio clip of astronaut Neil Armstrong’s words on the moon
- A written text discussing DNA’s advantages as a promising data storage method
- Random data to illustrate the applicability to encrypted or compressed data
The researchers plan to develop customized versions of DNAformer tailored to different needs. They emphasize that their technology is scalable and adaptable, meaning it can be optimized for large-scale data storage applications, meeting market demands and future DNA synthesis and sequencing advancements.
The study was supported by The European Research Council (ERC Grant, DNAStorage), The European Innovation Council (EIC Grant, Project DiDAX) and The Israel Science Foundation (ISF).
For the article in Nature machine intelligence click here .


